Inferno with binomial glm with "Wilkinson-Rogers" format and visreg package
Let do a binomial glm with cbind(success, failure)
First, I prepare the data:
factor <- rnorm(7, 10, 2)
dta <- data.frame(p=c(2, 7, 8, 9, 10, 18, 7),
n=c(20, 20, 20, 20, 20, 20, 20))
The error is because the first term of the formula is not understood correctly; it must be a simple term.
For example, here it does not work either:
g <- glm(cbind(p=dta$p, n=dta$n) ~ factor,
family = binomial(link = "logit"))
First, I prepare the data:
factor <- rnorm(7, 10, 2)
dta <- data.frame(p=c(2, 7, 8, 9, 10, 18, 7),
n=c(20, 20, 20, 20, 20, 20, 20))
dta cannot be used directly; it must be a matrix, not a data.frame:
g <- glm(dta ~ factor,
family = binomial(link = "logit"))
# Error in model.frame.default(formula = dta ~ factor, drop.unused.levels = TRUE) :
# type (list) incorrect pour la variable 'dta'
g <- glm(as.matrix(dta) ~ factor,
family = binomial(link = "logit"))
coef(g)
(Intercept) factor
-0.767901152 -0.006062598
Great, it works.
But if you try to plot the effects with visreg package, it produced an error:
library(visreg)
visreg(g, xvar ="factor")
# Error in dimnames(x) <- dn :
# la longueur de 'dimnames' [1] n'est pas égale à l'étendue du tableau
For example, here it does not work either:
g <- glm(cbind(p=dta$p, n=dta$n) ~ factor,
family = binomial(link = "logit"))
But here, it works:
n <- dta$n
p <- dta$p
g <- glm(cbind(p=p, n=n) ~ factor,
family = binomial(link = "logit"))
library(visreg)
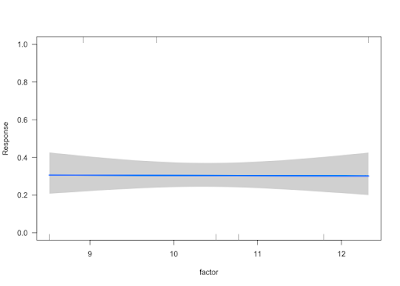
visreg(g, xvar ="factor", scale = "response", ylim=c(0, 1))
Thanks to Breheny, Patrick J <patrick-breheny@uiowa.edu> author of the visreg package for helping me to solve this problem.

Commentaires
Enregistrer un commentaire