Forking or not for parallel computing
In linux, forking is available when parallel computing is done but not in Windows. But what is the difference ?
Let do an exemple (code is below):
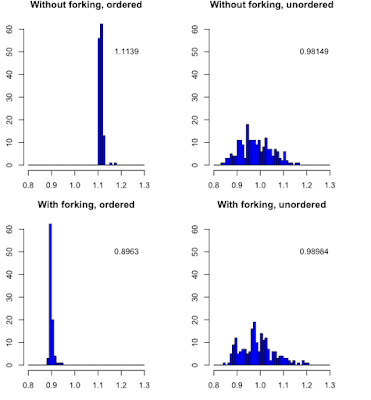
When the durations of the tasks are unordered, both algorithms are performing identically. However when task durations are ordered, forking is doing much better.
library(parallel)
l <- (1:32)/10/16.5
sum(l)
t0 <- system.time(lapply(l, FUN=function(x) {Sys.sleep(x)}))["elapsed"]
cl <- makeCluster(detectCores())
out1 <- NULL; for (i in 1:200) out1 <- c(out1, system.time(parLapplyLB(cl = cl, X = l, fun = function(x) {Sys.sleep(x)}))["elapsed"])
stopCluster(cl)
out2 <- NULL; for (i in 1:200) out2 <- c(out2, system.time(mclapply(l, mc.cores =detectCores(), FUN=function(x) {Sys.sleep(x)}))["elapsed"])
cl <- makeCluster(detectCores())
out3 <- NULL; for (i in 1:200) out3 <- c(out3, system.time(parLapplyLB(cl = cl, X = l[sample(32)], fun = function(x) {Sys.sleep(x)}))["elapsed"])
stopCluster(cl)
out4 <- NULL; for (i in 1:200) out4 <- c(out4, system.time(mclapply(l[sample(32)], mc.cores =detectCores(), FUN=function(x) {Sys.sleep(x)}))["elapsed"])
layout(mat = matrix(1:4, nrow = 2))
hist(out1, col="blue", breaks = seq(from=0.8, to=1.3, by=0.01), xlim=c(0.8, 1.3), ylim=c(0, 60),
main="Without forking, ordered")
text(x = 1.15, y=50, labels = format(mean(out1), digits = 5), pos=4)
hist(out2, col="blue", breaks = seq(from=0.8, to=1.3, by=0.01), xlim=c(0.8, 1.3), ylim=c(0, 60),
main="With forking, ordered")
text(x = 1.15, y=50, labels = format(mean(out2), digits = 5), pos=4)
hist(out3, col="blue", breaks = seq(from=0.8, to=1.3, by=0.01), xlim=c(0.8, 1.3), ylim=c(0, 60),
main="Without forking, unordered")
text(x = 1.15, y=50, labels = format(mean(out3), digits = 5), pos=4)
hist(out4, col="blue", breaks = seq(from=0.8, to=1.3, by=0.01), xlim=c(0.8, 1.3), ylim=c(0, 60),
main="With forking, unordered")
text(x = 1.15, y=50, labels = format(mean(out4), digits = 5), pos=4)
Let do an exemple (code is below):
When the durations of the tasks are unordered, both algorithms are performing identically. However when task durations are ordered, forking is doing much better.
library(parallel)
l <- (1:32)/10/16.5
sum(l)
t0 <- system.time(lapply(l, FUN=function(x) {Sys.sleep(x)}))["elapsed"]
cl <- makeCluster(detectCores())
out1 <- NULL; for (i in 1:200) out1 <- c(out1, system.time(parLapplyLB(cl = cl, X = l, fun = function(x) {Sys.sleep(x)}))["elapsed"])
stopCluster(cl)
out2 <- NULL; for (i in 1:200) out2 <- c(out2, system.time(mclapply(l, mc.cores =detectCores(), FUN=function(x) {Sys.sleep(x)}))["elapsed"])
cl <- makeCluster(detectCores())
out3 <- NULL; for (i in 1:200) out3 <- c(out3, system.time(parLapplyLB(cl = cl, X = l[sample(32)], fun = function(x) {Sys.sleep(x)}))["elapsed"])
stopCluster(cl)
out4 <- NULL; for (i in 1:200) out4 <- c(out4, system.time(mclapply(l[sample(32)], mc.cores =detectCores(), FUN=function(x) {Sys.sleep(x)}))["elapsed"])
layout(mat = matrix(1:4, nrow = 2))
hist(out1, col="blue", breaks = seq(from=0.8, to=1.3, by=0.01), xlim=c(0.8, 1.3), ylim=c(0, 60),
main="Without forking, ordered")
text(x = 1.15, y=50, labels = format(mean(out1), digits = 5), pos=4)
hist(out2, col="blue", breaks = seq(from=0.8, to=1.3, by=0.01), xlim=c(0.8, 1.3), ylim=c(0, 60),
main="With forking, ordered")
text(x = 1.15, y=50, labels = format(mean(out2), digits = 5), pos=4)
hist(out3, col="blue", breaks = seq(from=0.8, to=1.3, by=0.01), xlim=c(0.8, 1.3), ylim=c(0, 60),
main="Without forking, unordered")
text(x = 1.15, y=50, labels = format(mean(out3), digits = 5), pos=4)
hist(out4, col="blue", breaks = seq(from=0.8, to=1.3, by=0.01), xlim=c(0.8, 1.3), ylim=c(0, 60),
main="With forking, unordered")
text(x = 1.15, y=50, labels = format(mean(out4), digits = 5), pos=4)

Commentaires
Enregistrer un commentaire