Negative binomial and Poisson distributions
It is known that Poisson distribution is a special case of Negative binomial distribution. But how to parametrize NB to make it similar to Poisson in R ?
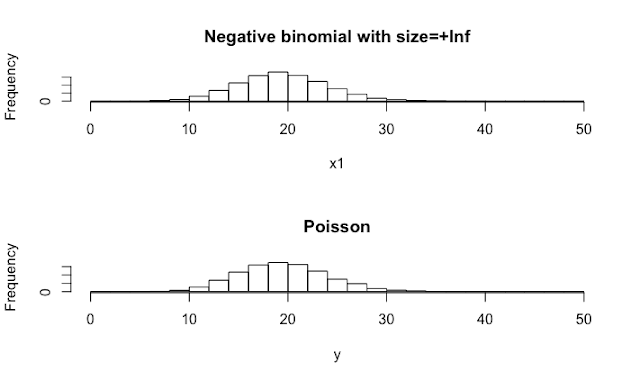
It is simple: rnbinom(n, mu=m, size=+Inf) is similar to rpois(n, lambda=m).
Try this to be sure !
layout(mat = 1:2)
x1 <- rnbinom(10000, mu=20, size =+Inf)
hist(x1, breaks=seq(from=0, to=50, by=2), main="Negative binomial with size=+Inf")
y <- rpois(10000, lambda=20)
hist(y, breaks=seq(from=0, to=50, by=2), main="Poisson")
Or this:
> dnbinom(x = 20, mu=20, size=+Inf)
[1] 0.08883532
> dpois(x=20, lambda=20)
[1] 0.08883532
Effect of size parameter, remember that size=+Inf is identical to Poisson distribution:
layout(mat = matrix(1:3, nrow= 1, byrow = TRUE))
x2 <- rnbinom(10000, mu=20, size =2)
hist(x2, breaks=seq(from=0, to=150, by=2), main="Negative binomial with size=2")
x2 <- rnbinom(10000, mu=20, size =10)
hist(x2, breaks=seq(from=0, to=150, by=2), main="Negative binomial with size=10")
x2 <- rnbinom(10000, mu=20, size =+Inf)
hist(x2, breaks=seq(from=0, to=150, by=2), main="Negative binomial with size=+Inf")
The better flexibility of the negative binomial distribution to fit counts can be seen also in my package phenology.
> library("phenology")
>
> gr <- add_phenology(add=Gratiot, previous = NULL, month_ref = 1, format="%d/%m/%Y", name="Cayenne2001")
Cayenne2001
Reference: 2001-01-01
>
> # fit the data using negative binomial distribution
>
> layout(mat = matrix(1:2, nrow= 1, byrow = TRUE))
>
> pfixed <- c(Flat=0)
> parg <- par_init(data=gr, parametersfixed = pfixed)
> parg3 <- MinBMinE_to_Min(parg)
>
> f <- fit_phenology(data=gr, parametersfit = parg3, parametersfixed = pfixed,
+ trace=0, silent=TRUE)
> outf <- plot(f, main="Negative binomial")
>
> # fit the data using Poisson distribution
>
> pfixed <- c(Flat=0, Theta=+Inf)
> parg <- par_init(data=gr, parametersfixed = pfixed)
> parg3 <- MinBMinE_to_Min(parg)
>
> f1 <- fit_phenology(data=gr, parametersfit = parg3, parametersfixed = pfixed,
+ trace=0, silent=TRUE)
> outf1 <- plot(f1, main="Poisson")
>
> # Compare both models
> compare_AIC(Negative.binomial=f, Poisson=f1)
[1] "The lowest AIC (1350.896) is for series Negative.binomial with Akaike weight=1.000"
AIC DeltaAIC Akaike_weight
Negative.binomial 1350.896 0.0000 1.000000e+00
Poisson 1741.075 390.1792 1.877852e-85
>
> # Compare the percentage of observations out of +/- 2 SD
> print("Proportion of observations out of +/- 2 SD for negative binomial distribution")
[1] "Proportion of observations out of +/- 2 SD for negative binomial distribution"
> sum((outf[[1]]$values[, "Obs"] > outf[[1]]$values[, "Theor+2SD"]) | (outf[[1]]$values[, "Obs"] < outf[[1]]$values[, "Theor-2SD"]), na.rm=TRUE) /
+ sum(!is.na(outf[[1]]$values[, "Obs"]))
[1] 0.04383562
>
> print("Proportion of observations out of +/- 2 SD for Poisson distribution")
[1] "Proportion of observations out of +/- 2 SD for Poisson distribution"
> sum((outf1[[1]]$values[, "Obs"] > outf1[[1]]$values[, "Theor+2SD"]) | (outf1[[1]]$values[, "Obs"] < outf1[[1]]$values[, "Theor-2SD"]), na.rm=TRUE) /
+ sum(!is.na(outf1[[1]]$values[, "Obs"]))
[1] 0.2082192
It is simple: rnbinom(n, mu=m, size=+Inf) is similar to rpois(n, lambda=m).
Try this to be sure !
layout(mat = 1:2)
x1 <- rnbinom(10000, mu=20, size =+Inf)
hist(x1, breaks=seq(from=0, to=50, by=2), main="Negative binomial with size=+Inf")
y <- rpois(10000, lambda=20)
hist(y, breaks=seq(from=0, to=50, by=2), main="Poisson")
Or this:
> dnbinom(x = 20, mu=20, size=+Inf)
[1] 0.08883532
> dpois(x=20, lambda=20)
[1] 0.08883532
Effect of size parameter, remember that size=+Inf is identical to Poisson distribution:
layout(mat = matrix(1:3, nrow= 1, byrow = TRUE))
x2 <- rnbinom(10000, mu=20, size =2)
hist(x2, breaks=seq(from=0, to=150, by=2), main="Negative binomial with size=2")
x2 <- rnbinom(10000, mu=20, size =10)
hist(x2, breaks=seq(from=0, to=150, by=2), main="Negative binomial with size=10")
x2 <- rnbinom(10000, mu=20, size =+Inf)
hist(x2, breaks=seq(from=0, to=150, by=2), main="Negative binomial with size=+Inf")
The better flexibility of the negative binomial distribution to fit counts can be seen also in my package phenology.
> library("phenology")
>
> gr <- add_phenology(add=Gratiot, previous = NULL, month_ref = 1, format="%d/%m/%Y", name="Cayenne2001")
Cayenne2001
Reference: 2001-01-01
>
> # fit the data using negative binomial distribution
>
> layout(mat = matrix(1:2, nrow= 1, byrow = TRUE))
>
> pfixed <- c(Flat=0)
> parg <- par_init(data=gr, parametersfixed = pfixed)
> parg3 <- MinBMinE_to_Min(parg)
>
> f <- fit_phenology(data=gr, parametersfit = parg3, parametersfixed = pfixed,
+ trace=0, silent=TRUE)
> outf <- plot(f, main="Negative binomial")
>
> # fit the data using Poisson distribution
>
> pfixed <- c(Flat=0, Theta=+Inf)
> parg <- par_init(data=gr, parametersfixed = pfixed)
> parg3 <- MinBMinE_to_Min(parg)
>
> f1 <- fit_phenology(data=gr, parametersfit = parg3, parametersfixed = pfixed,
+ trace=0, silent=TRUE)
> outf1 <- plot(f1, main="Poisson")
>
> # Compare both models
> compare_AIC(Negative.binomial=f, Poisson=f1)
[1] "The lowest AIC (1350.896) is for series Negative.binomial with Akaike weight=1.000"
AIC DeltaAIC Akaike_weight
Negative.binomial 1350.896 0.0000 1.000000e+00
Poisson 1741.075 390.1792 1.877852e-85
>
> # Compare the percentage of observations out of +/- 2 SD
> print("Proportion of observations out of +/- 2 SD for negative binomial distribution")
[1] "Proportion of observations out of +/- 2 SD for negative binomial distribution"
> sum((outf[[1]]$values[, "Obs"] > outf[[1]]$values[, "Theor+2SD"]) | (outf[[1]]$values[, "Obs"] < outf[[1]]$values[, "Theor-2SD"]), na.rm=TRUE) /
+ sum(!is.na(outf[[1]]$values[, "Obs"]))
[1] 0.04383562
>
> print("Proportion of observations out of +/- 2 SD for Poisson distribution")
[1] "Proportion of observations out of +/- 2 SD for Poisson distribution"
> sum((outf1[[1]]$values[, "Obs"] > outf1[[1]]$values[, "Theor+2SD"]) | (outf1[[1]]$values[, "Obs"] < outf1[[1]]$values[, "Theor-2SD"]), na.rm=TRUE) /
+ sum(!is.na(outf1[[1]]$values[, "Obs"]))
[1] 0.2082192



Commentaires
Enregistrer un commentaire